Defaults¶
Settings¶
Settings in p3, are defined in JSON format and can be loaded in through the -s or –settings command line flags. This document describes the settings available to default workflows of p3.
bids_query¶
Sets the bids query for the bidsselector workflow. The input to this settings key should be a dictionary containing the anat and func keys. See pybids docs for more information.
# This sets the anatomy images to be of modality 'anat' and type 'T1w'
# and the functional images to be modality 'func' and type 'rest'
{
'anat':{
'modality': 'anat',
'type':'T1w',
},
'func':{
'modality':'func',
'task':'rest'
}
}
func_reference_run¶
Selects the epi run to take the reference image from. It is 0 indexed so the first run loaded in to the func key would be 0.
func_reference_frame¶
Selects the epi reference frame to use. It is 0 indexed and taken from the whatever run was set to the func_reference_run.
anat_reference¶
Selects the anat to align to if multiple anat images in dataset. It is 0 indexed. Anatomy imagess are ordered from lowest session, lowest run to highest session, highest run. Leave as 0 if only 1 anat.
atlas¶
Sets the atlas align target. You can specify a path to an atlas here, or use one of p3’s built in templates. Currently the only availiable template in p3 is MNI152.nii.gz
avganats¶
True or False. Averages all anats in the dataset if multiple T1s. Set this to False if you only have 1 anatomy image or you will probably get an error!
field_map_correction¶
True or False. Sets whether the pipeline should run field map correction. You should have field maps in your dataset for this to work. p3 currently only supports gradient echo field maps.
slice_time_correction¶
True or False. Sets whether the functional images should be slice time corrected.
despiking¶
True of False. Sets whether epi images should be despiked.
run_recon_all¶
True or False. Sets whether pipeline should run recon-all (if you decide not to you should place your own p3_freesurfer data under output p3_freesurfer_output, where each folder is {NAME} in sub-{NAME} in the bids dataset)
num_threads¶
Sets the number of threads for all ANTs programs.
brain_radius¶
Sets the brain radius for FD calculations (in mm).
min_bpm¶
Sets the breathing rate for lower bound of the respiratory filter.
max_bpm¶
Sets the breathing rate for upper bound of the respiratory filter.
FD_threshold¶
FD threshold for creating tmask outputs.
FD_filtered_threshold¶
Filtered FD threshold for creating filtered tmask outputs.
workflows¶
Defines the workflows to import.
"workflows": [
"p3_bidsselector",
"p3_freesurfer",
"p3_skullstrip",
"p3_stcdespikemoco",
"p3_fieldmapcorrection",
"p3_alignanattoatlas",
"p3_alignfunctoanat",
"p3_alignfunctoatlas",
"p3_create_fs_masks"
]
See Creating New Workflows for more details.
connections¶
Defines the connections between workflows.
"connections": [
{
'source': 'p3_bidsselector', # the source workflow
'destination': 'p3_freesurfer', # the destination workflow
'links': [
[ # this sets the anat of p3_bidsselector to T1 of p3_freesurfer
'output.anat',
'input.T1'
],
[ # this sets the subject of p3_bidsselector to subject of p3_freesurfer
'output.subject',
'input.subject'
]
]
},
{
'source': 'p3_bidsselector', # the source workflow
'destination': 'p3_skullstrip', # the destination workflow
'links': [
[ # this sets the anat of p3_bidsselector to T1 of p3_skullstrip
'output.anat',
'input.T1'
]
]
},
...
]
See Creating New Workflows for more details.
sideload¶
A list of inputs to sideload a node.
"sideload": [
{
"workflow": "myworkflow1",
"node": "node1",
"input": ["field1", "value1"] # a single string sideload
},
{
"workflow": "myworkflow2",
"node": "node2",
"input": ["field2", ["value2_1","value2_2","value2_3"]] # an array sideload
}
]
See Sideloading for more details.
Outputs¶
Note
It should be noted that these are the outputs of the default workflows for the p3 pipeline. Changes to the workflow will not guarantee that you will have the same outputs as these or that they will be named the same.
graph¶
This folder contains the graph outputs of each subworkflow as well as the overall workflow (saved as p3.png).
freesurfer¶
This folder contains the freesurfer outputs. There is a skullstrip subfolder for storing the brainmask used in the default p3 skullstrip workflow. Freesurfer outputs for each subject are under each subject folder prefixed with sub-.
p3¶
This folder contains the main outputs of the p3 pipeline. Outputs that are generally used for further preprocessing stages are placed here. Subject outputs are separated into subfolders prefixed with sub-. The structure of each subject folder should be the following:
- atlas
Contains the atlas image resampled to the space of the functional reference image:
sub-(subject)/atlas/(atlas)_funcres.nii.gz # resampled atlas
- anat
Contains the processed anatomy image, and its resampled functional space version:
sub-(subject)/anat/(processed_anat).nii.gz # processed aligned anatomy image sub-(subject)/anat/(processed_anat)_funcres.nii.gz # resampled version
- func
This folder has an option session directory output if session are detected, prefixed with ses-. Aligned processed functional images are output here:
sub-(subject)/func/ses-(session)/(processed_func_image)_moco_atlas.nii.gz # aligned functional image sub-(subject)/func/ses-(session)/(func).1D # motion parameters sub-(subject)/func/ses-(session)/(func).FD # framewise displacement sub-(subject)/func/ses-(session)/(func).tmask # temporal mask sub-(subject)/func/ses-(session)/(funce)_filtered.1D # filtered motion parameters sub-(subject)/func/ses-(session)/(func)_filtered.FD # filtered framewise displacment sub-(subject)/func/ses-(session)/(func)_filtered.tmask # temporal Mask
p3_QC¶
This folder contains the QC outputs of the p3 pipeline. Outputs that are only useful for quality checking and are most likely not used for futher processing stages are placed here. All subfolders have a subject folder prefixed by sub-. The following subfolders are the following:
- alignfunctoanat
This contains QC outputs for the functional to anatomy alignment workflow:
sub-(subject)(processed_func)_reference_unwarped_skullstrip.nii.gz # skullstripped functional reference image sub-(subject)/(processed_func)_reference_unwarped_skullstrip_anat.nii.gz # aligned reference image sub-(subject)/(avg_anat)_skullstrip_corrected.nii.gz # skullstripped bias field corrected anatomy image
- bidsselector
This contains QC outputs from the bidsselector workflow. Despite it’s name this workflow also does the alignement of each anatomy image to each other and averages them together. The QC output here is for checking these alignments:
sub-(subject)/(anat).nii.gz # raw anatomy image sub-(subject)/(anat)_allineate.nii.gz # aligned anatomy image sub-(subject)/(anat)_avg.nii.gz # averaged anatomy image
- fieldmapcorrection
This contains QC outputs for the field map correction workflows:
sub-(subject)/(processed_func)_reference.nii.gz # functional reference image sub-(subject)/(processed_func)_reference_unwarped.nii.gz # unwarped functional reference image sub-(subject)/(processed_func).nii.gz # processed functional image sub-(subject)/(processed_func)_unwarped.nii.gz # unwarped functional image sub-(subject)/(processed_func)_unwarped_realign.nii.gz # realigned unwarped functional image
- skullstrip
This contains the QC outputs for the skullstrip workflow:
sub-(subject)/(anat)_avg.nii.gz # averaged anatomy image sub-(subject)/(anat)_avg_skullstrip.nii.gz # skullstripped anatomy image sub-(subject)/(anat)_avg_skullstrip_corrected.nii.gz # bias field corrected skullstripped anatomy image
- stcdespikemoco
This contains the QC outputs for the slice time corrected/despike/motion correction workflow:
sub-(subject)/(func_processed)_Warped.nii.gz # processed motion corrected functional image
fs_masks¶
This folder contains the freesurfer mask outputs. They are eroded by several levels denoted by the _erodeX suffix.
- aparc_aseg
- freesurfer segmentation volume
- cb
- cerbellum mask
- csf
- csf mask
- gm
- gray matter mask
- gmr
- gray matter ribbon
scn subcortical nuclei mask
- wm
- white matter mask
Workflows¶
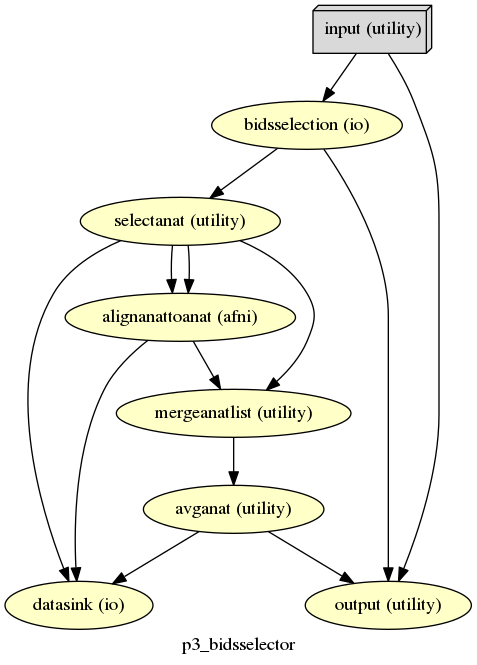
p3_bidsselector¶
description¶
The starting workflow of p3. Takes in a list of subjects from the ‘subject’ settings then processes the BIDS dataset for those subjects using the ‘bids_query’ setting. This workflow also averages the list of anatomical if the “avganat” setting is set.
output¶
anat: path to anatomical image func: list of paths to functional images subject: string of current subject being processed
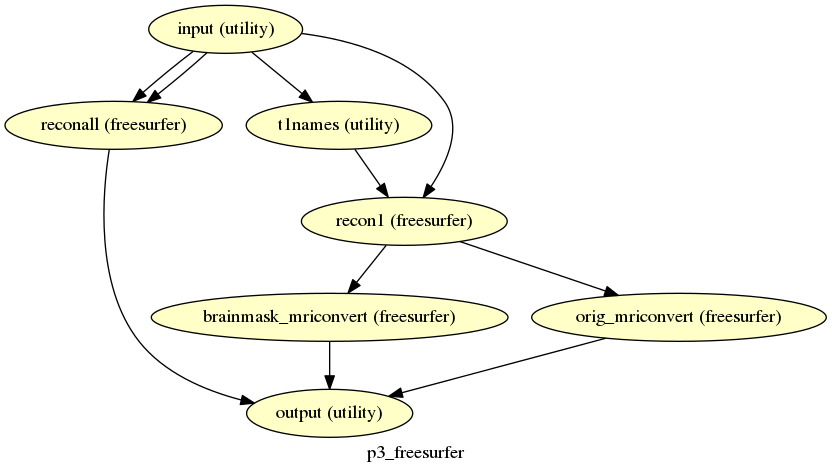
p3_freesurfer¶
description¶
Runs recon-all (and recon1 for the skullstrip).
input¶
T1: path to T1 image subject: string of current subject being processed
output¶
orig: freesurfer brainmasked T1 brainmask: freesurfer brainmask aparc_aseg: segmentation volume
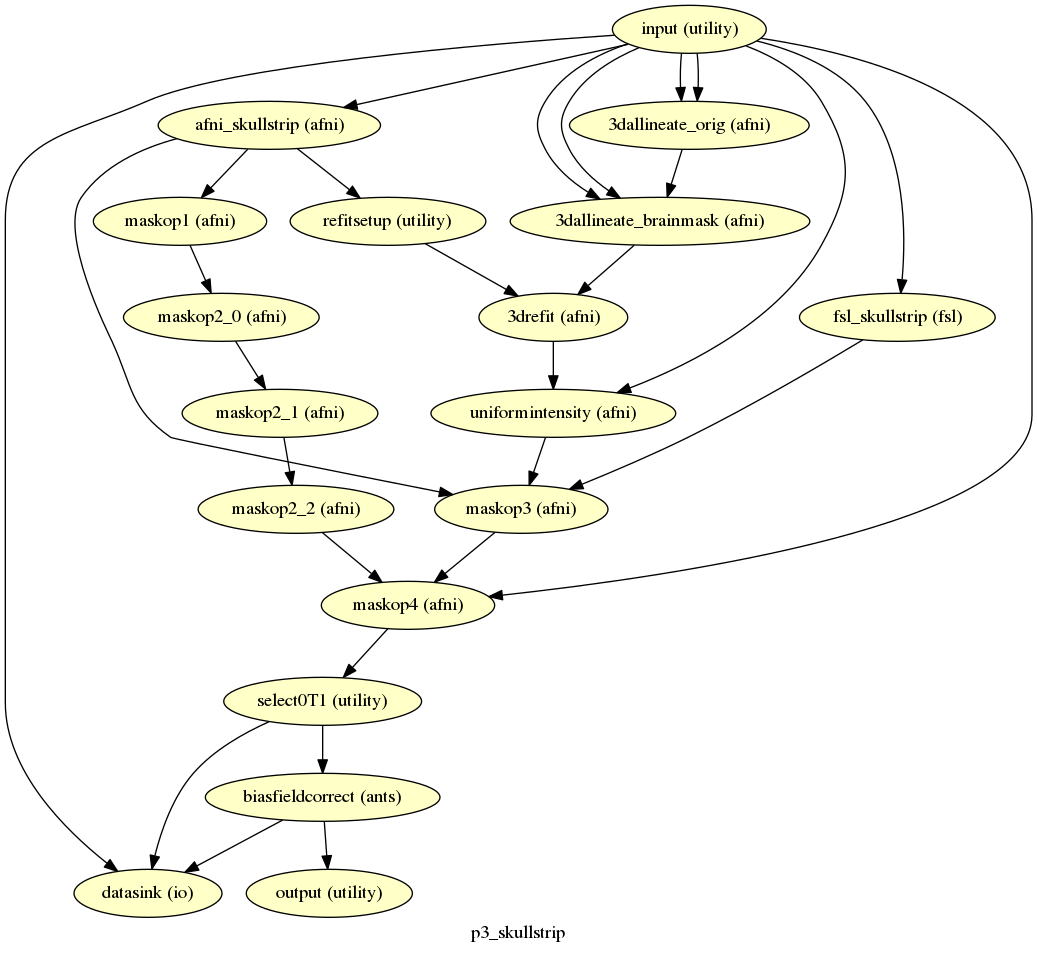
p3_skullstrip¶
description¶
Generates a skullstrip on the input T1. Uses Jonathan Power’s skullstrip algorithm that combines AFNI,FSL, and Freesurfer.
input¶
orig: freesurfer brainmasked T1 brainmask: freesurfer brainmask T1: path to T1 image
output¶
T1_skullstrip: path to skullstripped T1 image allineate_freesurfer2anat: 3dAllineate transfrom from freesurfer to anatomy image
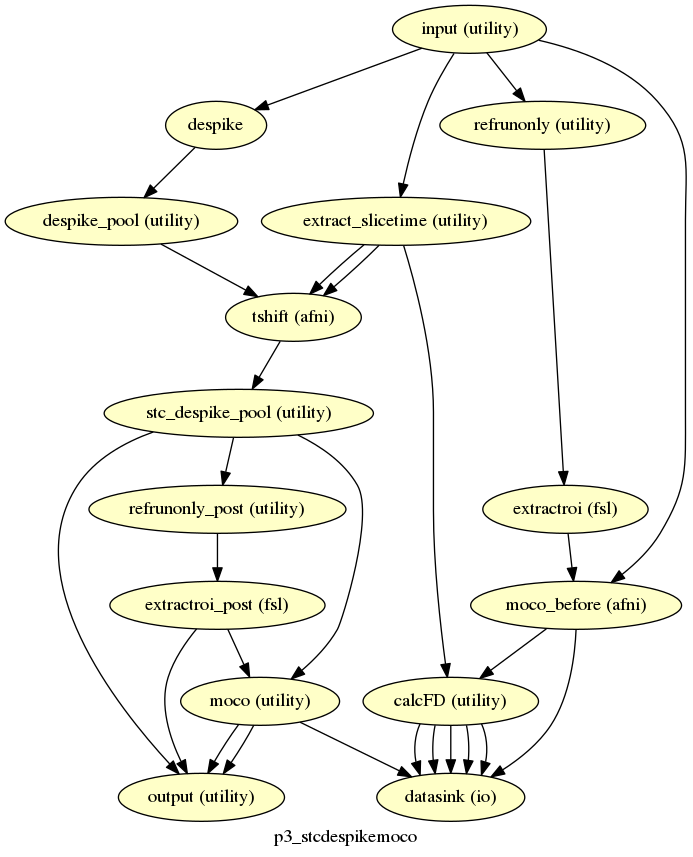
p3_stcdespikemoco¶
description¶
Does slice time correction, despiking, and motion correction. Produces the motion numbers, FDs and tmask.
input¶
func: list of paths to functional images
output¶
refimg: path to functional reference image func_stc_despike: list of paths to slice time corrected/despiked functional images warp_func_2_refimg: list of ANTs warp field transforms from functional to reference func_aligned: list of paths to aligned functional images
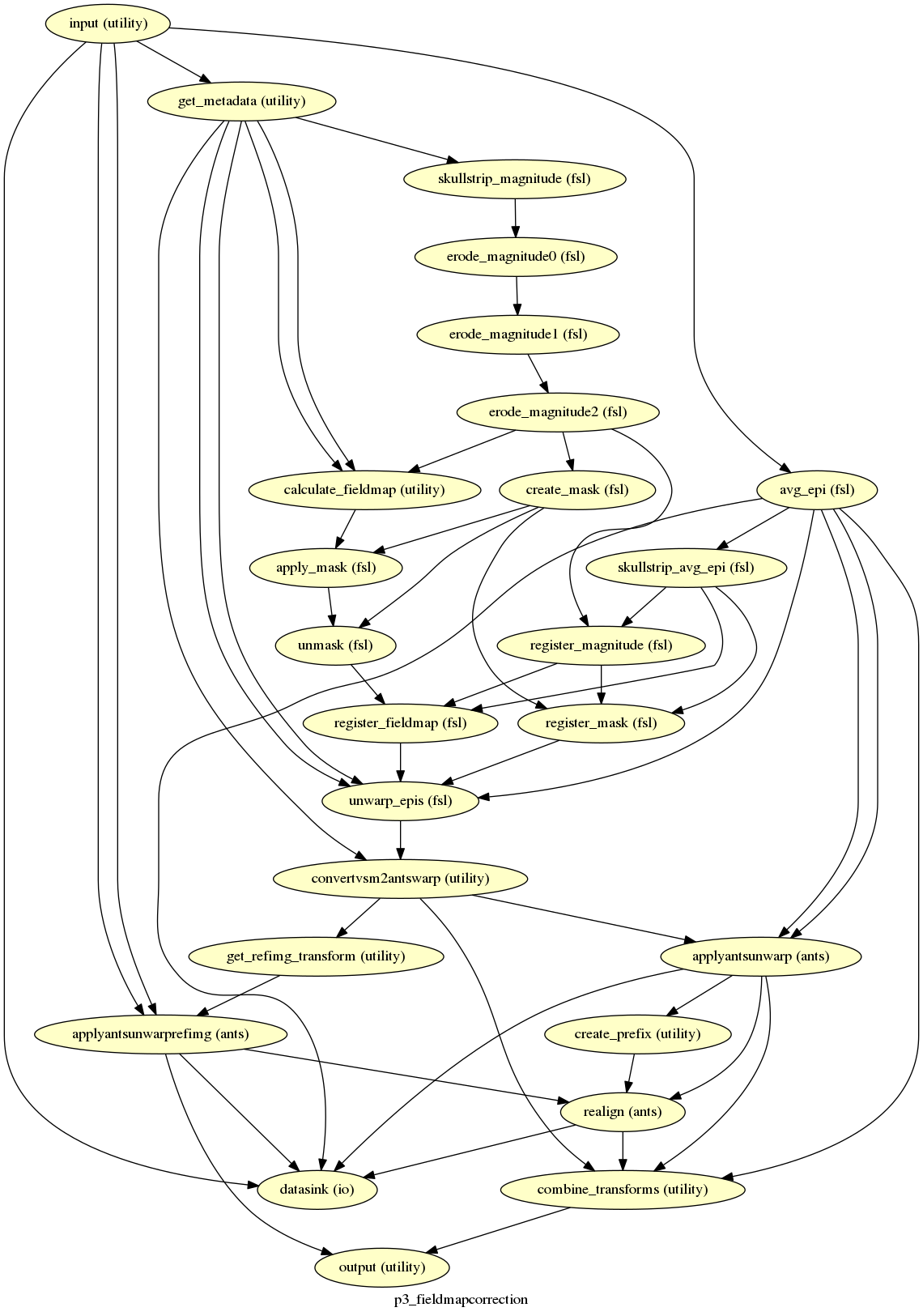
p3_fieldmapcorrection¶
description¶
Does FSL fieldmap correction on each functional image and then realigns the images back to the reference image.
input¶
func: list of paths to functional images refimg: path to functional reference image func_aligned: list of paths to aligned functional images
output¶
warp_fmc: list of ANTs warp field transforms for field map correction (aligned functional –> field map corrected functional) refimg: path to functional reference image (field map corrected)
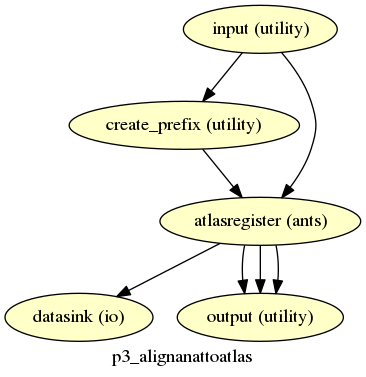
p3_alignanattoatlas¶
description¶
Aligns the anatomy image to the atlas.
input¶
T1_skullstrip: path to skullstripped T1 image
output¶
affine_anat_2_atlas: ANTs affine tranform from anatomical to atlas warp_anat_2_atlas: ANTs warp field transform from anatomical to atlas anat_atlas: atlas aligned anatomical image
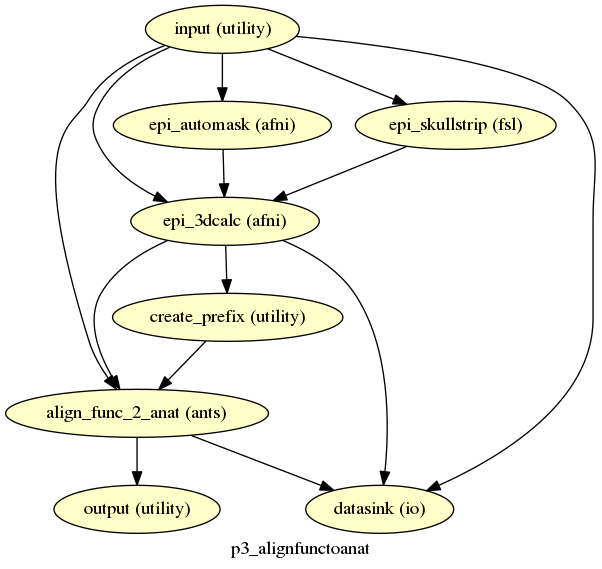
p3_alignfunctoanat¶
description¶
Aligns the functional reference to the anatomical image.
input¶
refimg: path to functional reference image (field map corrected if desired) T1_skullstrip: path to skullstripped T1 image
output¶
affine_func_2_anat: ANTs affine tranform from functional reference to anatomical warp_funct_2_anat: ANTs warp field transform from functional reference to anatomical
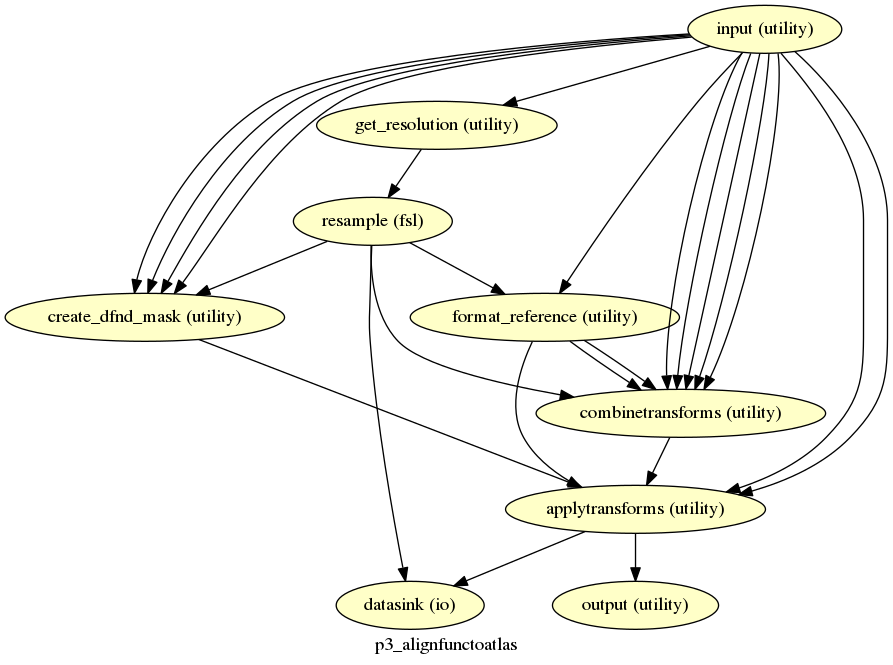
p3_alignfunctoatlas¶
description¶
Combines all the transforms and then aligns the functional images to the atlas.
input¶
func: list of paths to functional images func_stc_despike: list of paths to slice time corrected/despiked functional images warp_func_2_refimg: list of ANTs warp field transforms from functional to reference warp_fmc: list of ANTs warp field transforms for field map correction (aligned functional –> field map corrected functional) refimg: path to functional reference image (no field map correction) affine_func_2_anat: ANTs affine tranform from functional reference to anatomical warp_funct_2_anat: ANTs warp field transform from functional reference to anatomical affine_anat_2_atlas: ANTs affine tranform from anatomical to atlas warp_anat_2_atlas: ANTs warp field transform from anatomical to atlas
output¶
func_atlas: list of paths to atlas aligned functional images
p3_create_fs_masks¶
description¶
Create masks from the freesurfer outputs.
input¶
aparc_aseg: segmentation volume orig: freesurfer brainmasked T1 affine_anat_2_atlas: ANTs affine tranform from anatomical to atlas warp_anat_2_atlas: ANTs warp field transform from anatomical to atlas anat_atlas: atlas aligned anatomical image func_atlas: list of paths to atlas aligned functional images T1: path to T1 image